wellpaths: Generation of wells using general curves¶
Functionality for defining wells following curvilinear trajectories. By defining each well trajectory as a series of points, this module can combine multiple trajectories and determine which cells are penetrated by the well path. The trajectories can be interpolated using any of Matlab’s built-in routines (splines, piecewise linear functions, etc).
-
Contents¶ UTILS
- Files
- combineWellPaths - Combine multiple simple paths into a full tree findWellPathCells - Convert well path to the intersected cells getWellFromPath - Convert well path to MRST well. makeSingleWellpath - Create well path from points (and optional connectivity and active flag) plotWellPath - Plot a well path refineSpline - Refine a curve to higher resolution using spline interpolation
-
combineWellPaths(wellpaths)¶ Combine multiple simple paths into a full tree
Synopsis:
wellpath = combineWellPaths({wp1, wp2, wp3});
Description:
Given multiple simple well paths this function assembles a single well tree from the inputs. For this to work we assume that:
- The paths are ordered by depth in the tree. This is not the
vertical depth, but rather that a path will always be connected to one of the curves preceding it in the list. - Paths (aside from the first one) always start with a point that also exists in one of the preceding paths. This is used to connect the paths.
Parameters: wellpaths – Cell array of simple well paths to be assembled together. A simple well path is assumed to contain a single list of points (i.e. it will only represent a line segment). Returns: wellpath – Composite wellpath made from the simple wellpaths. The topology will be tree-like in nature. See also
-
findWellPathCells(G, wellpath, varargin)¶ Convert well path to the intersected cells
Synopsis:
cells = findWellPaths(G, wellpath);
Description:
By creating a triangulation and mapping perforations to the closest cells, this routine realizes the continuous well path into discrete cells, making it possible to build simulation wells from it.
Parameters: - G – The grid structure we want to realize the wells on.
- wellpath – Well path. See “makeSingleWellpath” for spec.
Keyword Arguments: - interpType – The type of interpolation used to extend the curve between points. Supports the same types as MATLAB builtin interp1. Default: Spline.
- triangulation – The triangulation (typically from delaunayTriangulation) used to determine proximity in the grid.
- refinement – Refinement number used to further refine the well curve before computing which cells it intersect. Default: 100.
Returns: - cells – The list of cells the well intersects.
- segment – Segment indicator for each cell, indicating which wellpath segment produced that specific completion. If multiple choices are possible, the segment which comes first in wellpath.points is used.
- ptsind – Point indicator, indicating which point in the segment was the closest to a given cell.
- DT – Triangulation used to produce the results.
See also
-
getWellFromPath(W0, G, rock, wellpath, varargin)¶ Convert well path to MRST well.
Synopsis:
W = getWellFromPath(W, G, rock, wellpath);
Description:
This routine converts a well path (representing curves and points) into a well (represented by cells and connectivity).
Parameters: - W0 – Well array to be extended with the new well.
- G – The grid the well is to be placed in.
- rock – Rock structure which defines permeability and porosity.
- wellpath –
- Well path as procued by makeSingleWellPath or
- combineWellPaths.
OPTIONAL PARAMETERS:
This function calls addWell. Any keyword arguments are passed onto addWell.
Returns: W – Updated wells
Note
Currently no special effort is made to ensure correct well indices for the well.
SEE ALSO:
-
makeSingleWellpath(pts, conn, active)¶ Create well path from points (and optional connectivity and active flag)
Synopsis:
wellpath = makeSingleWellpath(pts); wellpath = makeSingleWellpath(pts, conn) wellpath = makeSingleWellpath(pts, conn, active)
Description:
Create a well path from lists of points. The resulting structure will be a single struct with fields:
- points:
Cell array, each consisting of N x Dim arrays of n points. Each entry contains the points for one segment that are assumed to be connected as a line according to their ordering. The first entry is assumed to be closest to the starting point of the well (closest here means along the well bore).
- connectivity:
Array of size M x 2 where M is the number of entries in the .points cell array. If entry number 5 of connectivity is [2 7] it means that segment number 5 is connected to segment 2, at point number 7 of segment 2’s internal ordering. In effect, segment 5 branches off from segment 2 from the coordinate .points{2}(7, :).
- active:
Cell array, containing active flags for the segments between points. If points{i} contains N x dim entries, active{i} should contain (N-1) x 1 entries, indicating if the subsegments are active.
If points is of size 6 x 3 and active looks like this: [1; % 1 -> 2
1; % 2 -> 3 0; % 3 -> 4 1; % 4 -> 5 1] % 5 -> 6
it means that the part of the segment will be disabled from point 3 to point 4.
Parameters: pts – - Maps directly to points (see above). If pts is a numeric
- array, it will be interpreted as a cell array with a single entry.
connectivity - Maps directly into connectivity.
active - Maps directly into active.
Returns: wellpath – Wellpath suitable for plotting or producing well completions. See also
-
plotWellPath(wellpaths, varargin)¶ Plot a well path
Synopsis:
plotWellPath(wellpath) h = plotWellPath(wellpath, 'color', 'r')
Description:
Plots a given well path, using colors and showing control points along the curve.
Parameters: wellpath – Well path to be plotted. See makeSingleWellpath.
Keyword Arguments: - interpType – Interpolation type used. Same possible values as for MATLAB builtin interp1. Default: Spline.
- LineWidth – Line width of curve used to draw wellpath segments.
- MarkerColor – Used to colorize the control points.
- Color – Color of the line segments themselves.
- Refinement – The path is refined using interpolation to produce nice curves. Entering a positive number here will refine the curve by a number of points. Interpreted as a multiplicative factor.
Returns: h – Two-column array of handles. The first column contains handles for the line segments and the second for the control point markers.
See also
-
refineSpline(points, n_refine, interpType)¶ Refine a curve to higher resolution using spline interpolation
Synopsis:
pts = refineSpline(points, 10, 'spline'); pts = refineSpline(points, 5);
Description:
Refine a given curve given as a array of points into
Parameters: - points – A npts x dim array of points giving the curve to be refined. Implicitly assumed to be ordered.
- n_refine – The refinement factor. If the original entries in points contained n points, the output will have n_refine*n total points.
- interpType – Type of interpolation. Supports the same values as the fourth argument to MATLABs interp1 function. If omitted, it defaults to ‘spline’.
Returns: - pts – Refined points.
- v – Parametrization of the new points. Continuous values from 1 to npts indicating how far along interpolated values are on the original trajectory.
Examples¶
Define a model¶
Generated from wellTrajectoryExample.m
mrstModule add ad-core ad-blackoil diagnostics wellpaths
km = kilo*meter;
pdims = [5, 4, 0.01]'*km;
% dims = [50, 40, 10];
dims = [25, 20, 5];
G = cartGrid(dims, pdims);
G = computeGeometry(G);
Define a forked well¶
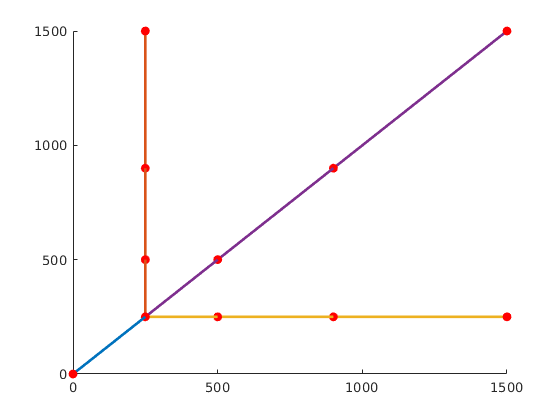
We define four curves, with some common points to combine them into a single well path.
p1 = [0.25, 0.5, 0.9, 1.5]'*km;
n = numel(p1);
z = [.25, 0.5, 0.75, .80]'*pdims(3);
a = [p1(1)*ones(n, 1), p1];
b = [p1, p1(1)*ones(n, 1)];
c = [p1, p1];
origin = [0, 0, 0; a(1, :), z(1)];
% First foru well paths.
wellpath0 = makeSingleWellpath(origin);
wellpath1 = makeSingleWellpath([a, z]);
wellpath2 = makeSingleWellpath([b, z]);
wellpath3 = makeSingleWellpath([c, z]);
% Combine into single well path
wellpath_fork = combineWellPaths({wellpath0, wellpath1, wellpath2, wellpath3});
% Plotting
clf;
plotWellPath(wellpath_fork);
Make a comb-like well¶
x = [3, 3.5, 4, 4.5]'*km;
y = [3.5, 3, 2.5, 2]'*km;
y0 = [3.75*km; y];
x0 = x(end)*ones(numel(y)+1, 1);
dz = 1/numel(y);
z0 = (0:dz:1)'*pdims(3);
wp0 = [x0, y0, z0];
n0 = numel(x0);
wellpaths = cell(5, 1);
wellpaths{1} = makeSingleWellpath(wp0);
for i = 1:4
n = numel(x);
wp = [x, repmat(y(i), n, 1), repmat(z0(i+1), n, 1)];
wellpaths{i+1} = makeSingleWellpath(wp(end:-1:1, :));
end
wellpath_comb = combineWellPaths(wellpaths);
Determine the cells¶
[cells_fork, segInd_fork, ~, ~, DT] = findWellPathCells(G, wellpath_fork);
[cells_comb, segInd_comb] = findWellPathCells(G, wellpath_comb, 'triangulation', DT);
Plot the well trajectories¶
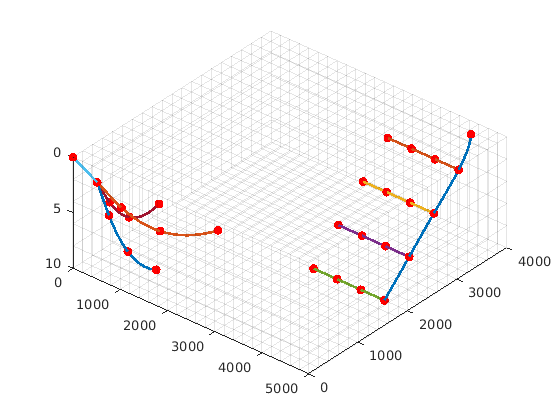
clf;
plotWellPath(wellpath_comb);
plotWellPath(wellpath_fork);
plotGrid(G, 'facec', 'none', 'edgea', .1)
view(40, 56)
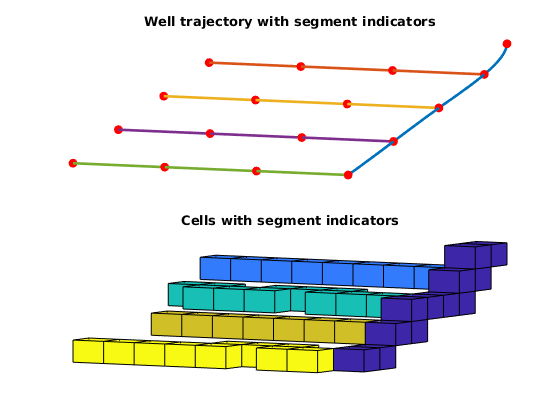
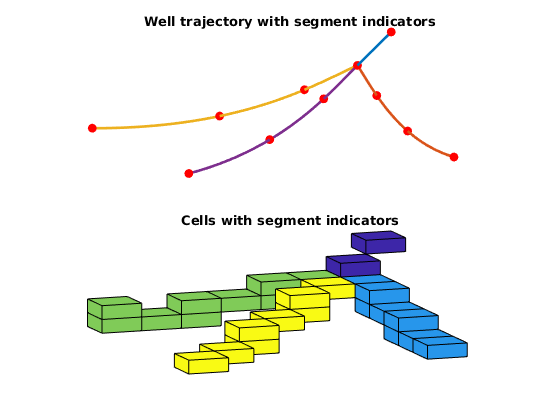
Plot wells individually + realized cell perforations¶
close all
for i = 1:2
if i == 1
wp = wellpath_comb;
si = segInd_comb;
c = cells_comb;
v = [30, 12];
else
wp = wellpath_fork;
si = segInd_fork;
c = cells_fork;
v = [160, 40];
end
figure;
subplot(2, 1, 1)
plotWellPath(wp);
view(v);
title('Well trajectory with segment indicators')
axis tight off
subplot(2, 1, 2)
plotCellData(G, si, c)
view(v);
title('Cells with segment indicators')
axis tight off
end
Set up actual simulation wells from the well paths¶
Initial reservoir state
initSat = [.1, .9];
state = initResSol(G, 200*barsa, initSat);
% Rock
rock = makeRock(G, 300*milli*darcy, 0.5);
time = 10*year;
rate = .25*sum(poreVolume(G, rock))/time;
segInd = cell(2, 1);
W = [];
[W, segInd{1}] = getWellFromPath(W, G, rock, wellpath_fork, ...
'comp_i', [1 0], 'val', 300*barsa, 'type', 'bhp', 'sign', -1, 'Name', 'Producer');
[W, segInd{2}] = getWellFromPath(W, G, rock, wellpath_comb,...
'comp_i', [1 0], 'val', rate, 'type', 'rate', 'sign', 1, 'Name', 'Injector');
Set up simulation model¶
mrstModule add ad-core ad-blackoil ad-props
fluid = initSimpleADIFluid('rho', [1000, 700, 100]*kilogram/meter^3, ...
'mu', [1, 10 1]*centi*poise);
model = TwoPhaseOilWaterModel(G, rock, fluid);
model.extraStateOutput = true;
model.extraWellSolOutput = true;
Build a schedule¶
n = 50;
dt = time/n;
timesteps = repmat(dt, n, 1);
schedule = simpleSchedule(timesteps, 'W', W);
[ws, states] = simulateScheduleAD(state, model, schedule);
Solving timestep 01/50: -> 73 Days
Solving timestep 02/50: 73 Days -> 146 Days
Solving timestep 03/50: 146 Days -> 219 Days
Solving timestep 04/50: 219 Days -> 292 Days
Solving timestep 05/50: 292 Days -> 1 Year
Solving timestep 06/50: 1 Year -> 1 Year, 73 Days
Solving timestep 07/50: 1 Year, 73 Days -> 1 Year, 146 Days
Solving timestep 08/50: 1 Year, 146 Days -> 1 Year, 219 Days
...
Plot well curves + reservoir properties¶
mrstModule add mrst-gui
figure;
plotToolbar(G, states);
axis tight off
view(40, 56);
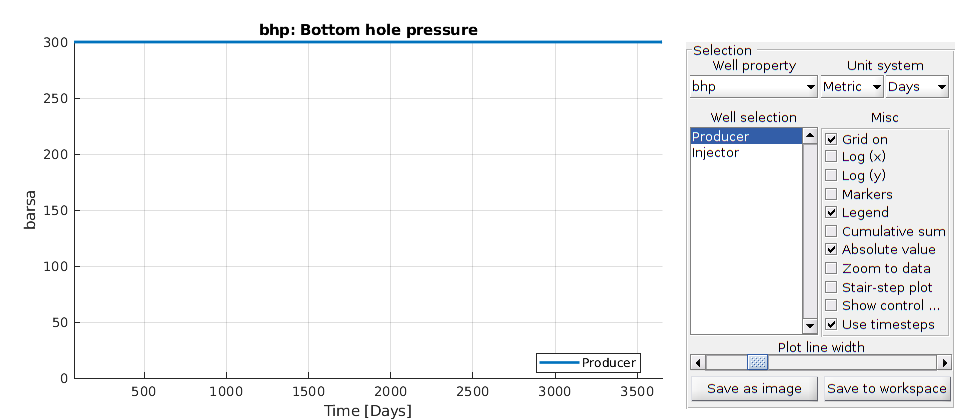
T = cumsum(schedule.step.val);
plotWellSols(ws, T)
Plot the saturation front¶
close all
plotWellPath(wellpath_comb);
plotWellPath(wellpath_fork);
plotGrid(G, 'facec', 'none', 'edgea', .2);
view(-60, 60);
h = nan;
for i = 1:numel(states)
if ishandle(h); delete(h); end
s = states{i}.s(:, 1);
h = plotCellData(G, s, s > 0.2, 'edgea', .2);
title(formatTimeRange(T(i)));
pause(0.25)
end
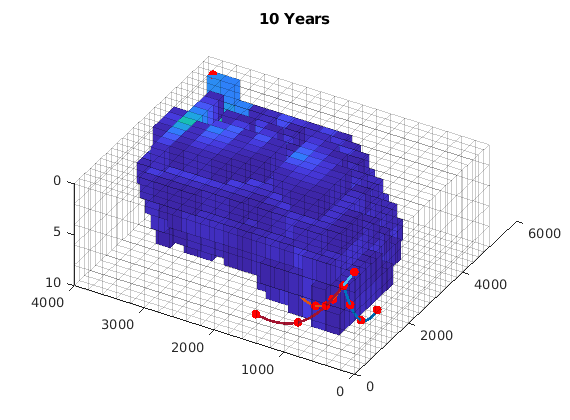
Apply some flow diagnostics…¶
diagstate = states{end};
[state_split, Wc] = expandWellCompletions(diagstate, W, segInd);
D = computeTOFandTracer(state_split, G, rock, 'wells', Wc);
WP = computeWellPairs(state_split, G, rock, Wc, D);
